CD Bioparticles offers custom services to stimuli-responsive controlled-release nanocarriers using advanced techniques. Our experienced scientists have created a comprehensive platform to design the enzyme-responsive controlled-release nanocarriers in order to improve the capability of controlled-release drug delivery system.
Enzymes underpin physiological function and exhibit dysregulation in many disease-associated microenvironments and aberrant cell processes. Exploiting altered enzyme activity and expression for site-specific enzyme-triggered drug release is tremendously promising. As major biocatalytic substances, enzymes are involved in many biochemical reactions within the body. For example, esterases catalyze ester hydrolytic reactions; glycosidases catalyze the hydrolysis of glycosidic bonds; peptidases catalyze the hydrolytic breakdown of proteins into amino acids; and reductases catalyze hydrogenation reactions. In addition, enzymes contribute to the biodegradation of biomacromolecules and some specific enzymes are over-expressed in cancer cells, indicating great potential for developing enzyme-sensitive controlled drug delivery systems.
In enzyme-responsive controlled-release nanocarriers, there are several methods to come true the gating. Firstly, the substrate is used the gatekeeper, and it can block the hole of mesoporous silica nanoparticles. Upon exposure to corresponding enzyme, gates were removed to release drug molecules. Secondly, drugs are conjugated to the polymer scaffold by some special bonds. After specific cell uptake, these bonds are broken by corresponding enzyme, and then the drug molecules can be released from the polymer. Thirdly, the nanomaterials can be rendered enzyme-responsive by containing moieties in their main chain or side groups which can be cleaved by enzyme. Upon exposure to corresponding enzyme, the nanomaterials can be degraded, and the drug is releasing.
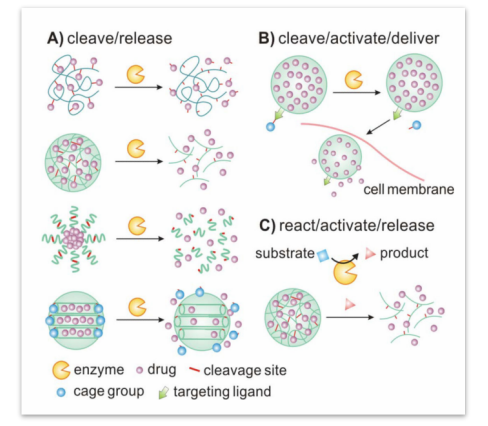
Figure 1. A schematic illustrating typical implementations of enzyme-responsive nanomaterials for controlled drug delivery. (Quanyin Hu, et al. Nanoscale. 2014, 6(21): 12273–12286.)
Enzyme-responsive drug delivery system has been witnessed a rapid development by exhibiting tremendous therapeutic and detection potency for cancer or other diseases at both research and clinical levels. By overcoming the limitations and drawbacks of conventional drug delivery, they offer unprecedented control over spatiotemporal drug release and delivery profiles leading to superior in vitro and/or in vivo theranostic efficiency. For example, active tumor targeting nanoparticles integrated with site-specific enzyme-triggered moieties can significantly achieve enhanced accumulation at the tumor site, reduced uptake by non-targeted tissue, as well as site-specific controlled drug release without a compromise in targeting efficiency and specificity. Oxidoreductases have been widely exploited to serve as a promising target for drug delivery systems or biosensing due to their fundamental role in oxidative environments which are generated by many diseases including diabetes and cancer. As many diseases are characterized by imbalances in the expression and activity of specific enzymes in the diseased tissue, dysregulated enzymes can be exploited to permit the selective activation of advanced drug delivery platforms.
Quotations and Ordering

References:
1. Quanyin H, et al. Enzyme-responsive nanomaterials for controlled drug delivery. Nanoscale. 2014, 6(21): 12273–12286.
2. Chendi D, et al. Recent advances in stimuli-responsive release function drug delivery systems for tumor treatment. Molecules. 2016, 21, 1715.
3. Chun-Ling Z., et al. Cell microenvironment stimuli-responsive controlled-release delivery systems based on mesoporous silica nanoparticles. Journal of food and drug analysis. 2014, 22: 18-28.
1. Download the template.
2. Enter product information on the template (maximum number of products: 200).
3. Load the file using selector below.
1. Download the template.
2. Enter product information on the template (maximum number of products: 200).
3. Load the file using selector below.